Research
Microbiome transmission in mothers and infants
At birth, the infant gut represents a “blank sheet”, which the microbes readily colonize. But where are the colonizing microbes coming from? To address this question, I started to investigate the most intimate relationship that the infant has: the one with the mother.

Ferretti and Allert et al. 2025 - Nature Communications
Ideally, the maternal breastmilk represents the sole source of nutrition for the infant in the first semester of life. Yet, milk represents less than 0.6% of all metagenomic samples publicly available. Little is known about the strain-level milk microbiome composition and stability, and its relevance for the infant’s health. In this recent study, I investigated the species- and strain-level composition of the breastmilk, its functional potential, and antimicrobial resistance in relation to the infant’s gut microbiome. I found that the breast milk microbiome directly contributes to the assembly of the infant gut microbiome, providing both commensal species (like Bifidobacteria) and pathobionts. You can find more info on the results of this study in the official press release here.
Relevance of the study: High-resolution characterization of the milk microbiome is a crucial step towards understanding its role in the assembly, development and stability of the infant gut microbiome, with the ultimate goal of improving health outcomes and current best practices.
Ferretti et al. 2018 - Cell Host & Microbe
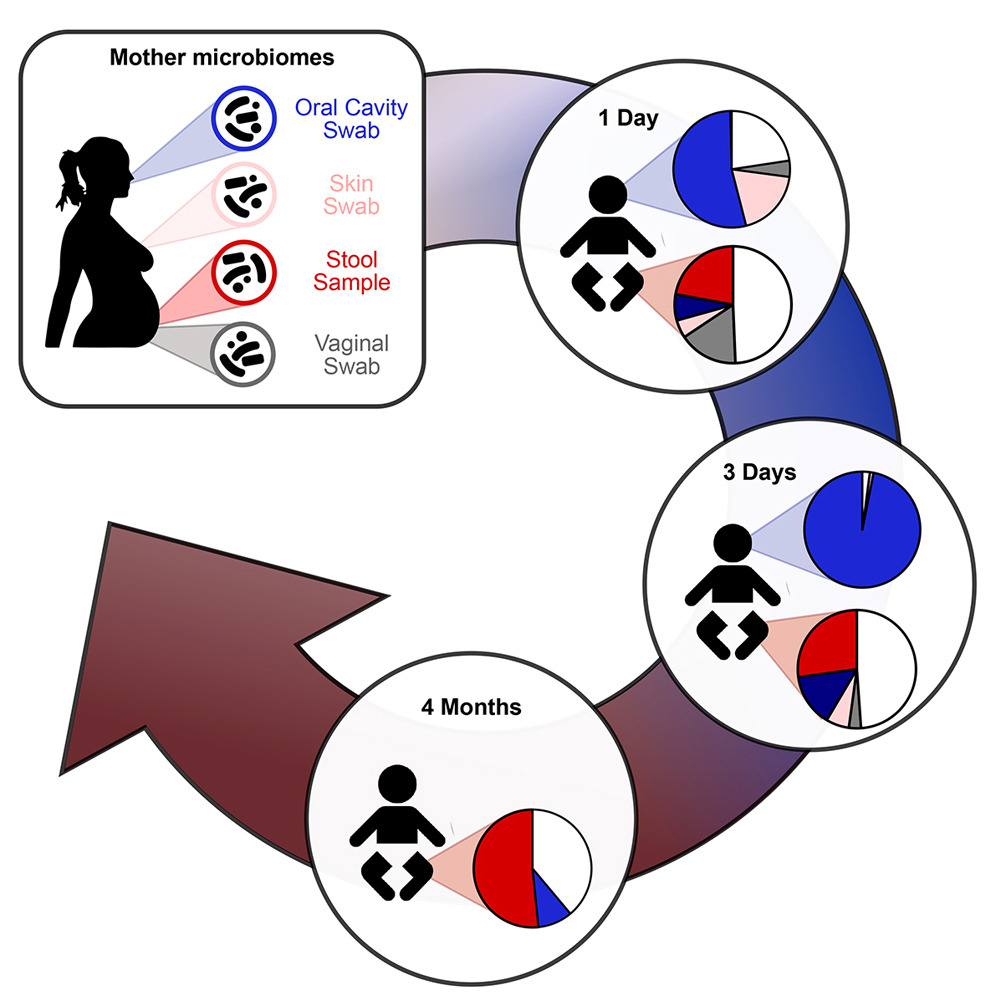
Here, I investigated how the maternal gut, skin, oral and vaginal microbiomes seed and shape the infant microbiome in the first semester of life. This study represented the first extensive charcaterization of mother-infant strain sharing across multiple body sites. For this project I collected 2,000+ samples over the course of 3 years, and I worked in close contact with the neonatologists and OB/GYNs at the Santa Chiara Hospital in Trento. This project sparked my interest in microbiome research, and I enjoyed every bit of it.
Relevance of the study: Understanding the routes of microbial transmission from the mother to her baby helps defining better healthcare policies, and strenghtens the importance of practices part of the Baby Friendly Hospital Initiative, such as skin-to-skin contact and rooming-in.
Microbiome transmission in social groups
Chege, Ferretti et al. 2025 - Animal Microbiome
Studying microbial transmission dynamics with enough high spatial and temporal resolution remains extremely challenging in human populations. For this reason, we are studying how bacteria and eukaryotes are transmitted in wild animal groups. I recently co-authored a study that identified social group membership as important factor in shaping the eukaryotes diversity and composition in the gut microbiome of wild baboons. Ongoing projects are currently investigating microbial strain sharing and antimicrobial resistance gene transmission within a unique dataset of wild baboons, representing the largest metagenomic survey of a wild animal population to date. Stay tuned for updates!
Defining, identifying and interpreting microbial strains
Van Rossum, Ferretti and Maistrenko 2020 - Nature Reviews Microbiology
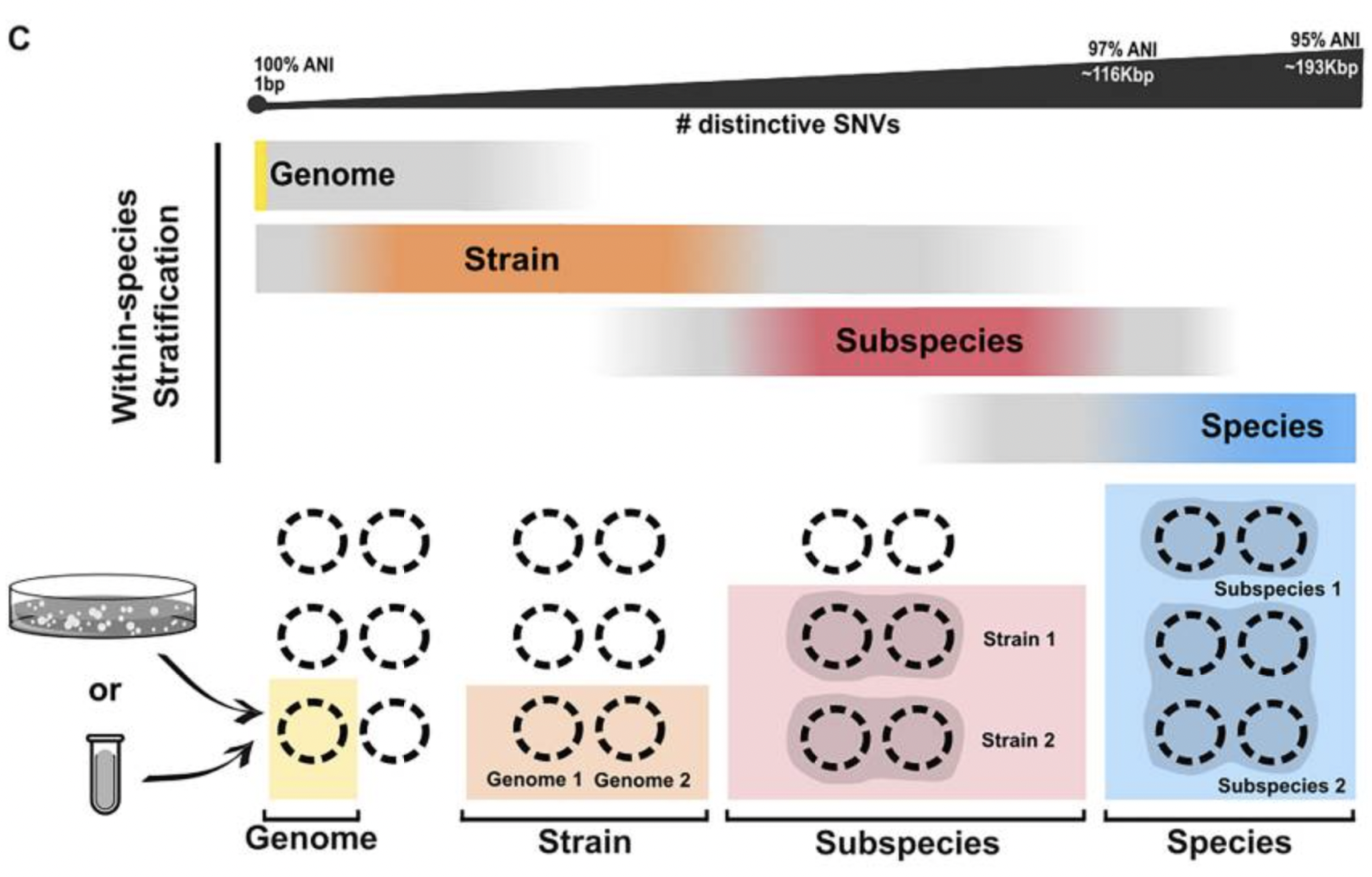
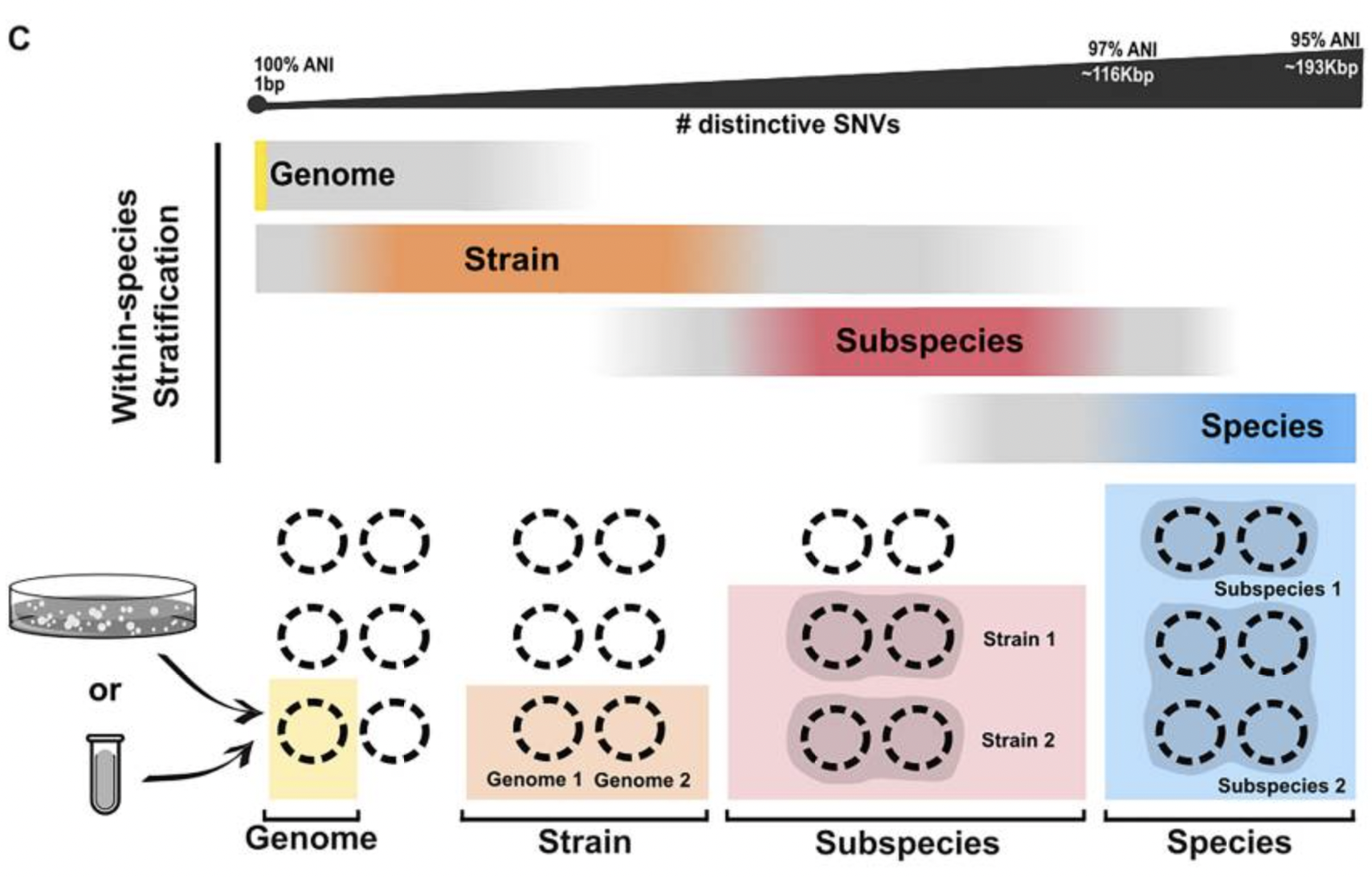
Most studies look at species-level composition, but often the presence (or absence) of a single gene, or a set of genes, can make the difference between health and disease. Hence, the importance of diving into the strain-level world. In this work, I explored (with Thea and Oleksandr) the mechanisms that drive variability within bacterial species and the challenges in stratifying such variability, providing harmonized terminology and the associated scope of applicability. This Review, which took >1 year to write and gave us all a fair amount of headaches, touches important aspects of microbial ecology that are widely applicable (from human microbiome, to animals, to marine microbial ecosystems).
Relevance of the study: Our ability to reproduce results and compare them among studies depends on common terminology (e.g. what is a strain?) and agreement on gold standards (e.g. when are MAGs good enough to be used in strain-level analyses?). Knowing the importance of strain-level analysis, its challenges, and the limitations of the available tools guides the researchers towards better study design, analysis and costs allocation.
Resource Development and Meta-Analyses on Large-Scale Metagenomic data
The biogeography of C. difficile: from CDI to healthy infants
C. difficile is one the most urgent threats in hospital-acquired infections. During my PhD, I leveraged public metagenomic data to investigate the composition and stability of the gut microbiome in presence of C. difficile. I found that patients diagnosed with C. difficile infection (CDI) had a suprisingly low carriage of C. difficile (only 30%). In these patients, I instead found elevated carriage of other pathogens known to be able to induce CDI-like symptomatology, suggesting CDI over-diagnosis in the evaluated studies.
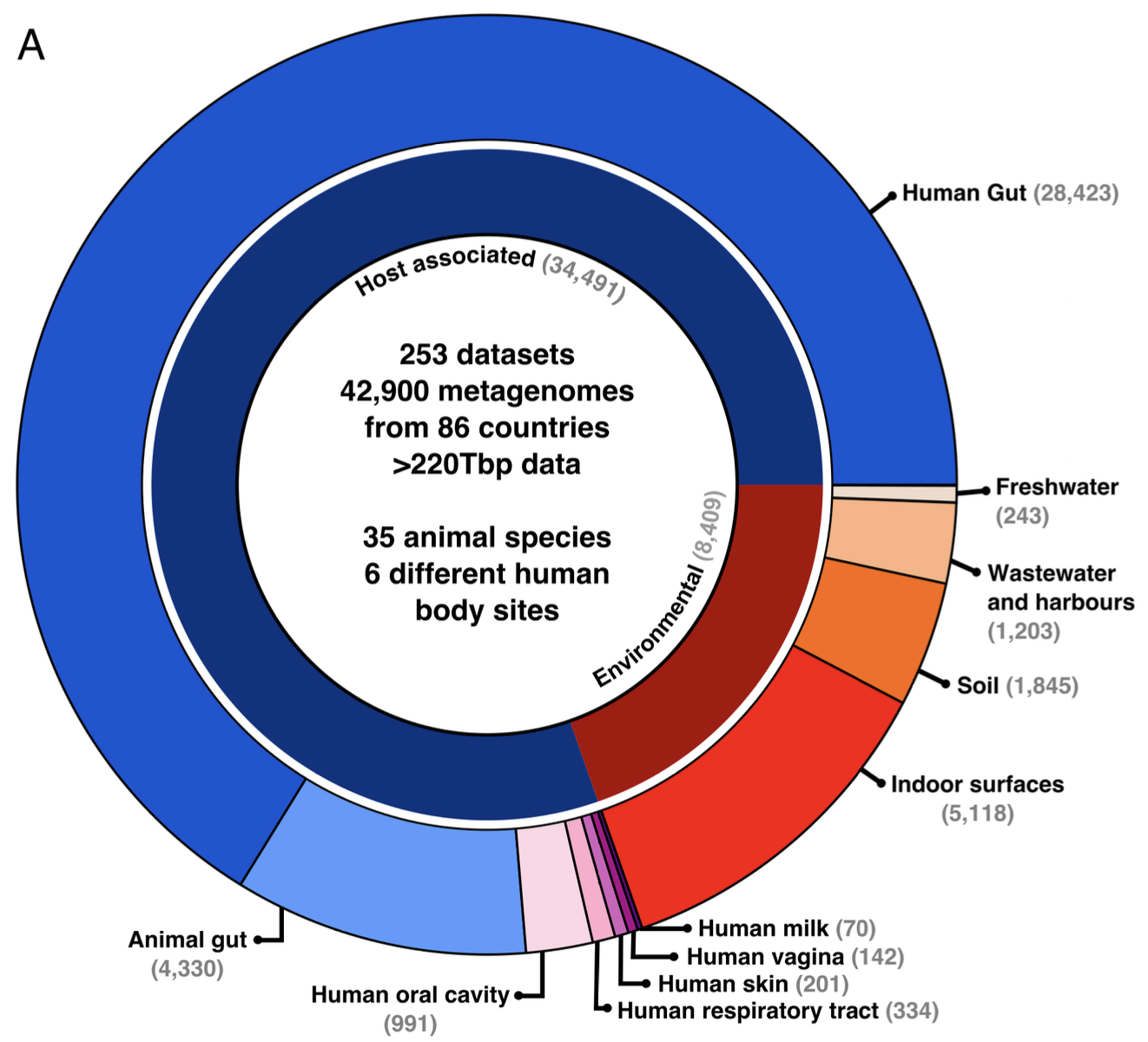
I then included in my meta-analysis healthy subjects of all ages, animal hosts and environmental samples. I assembled a collection of 42,900 metagenomic samples, curated their associated metadata (yes was a lot of work!), and then leveraged this dataset to investigate the global biogeography and genetic variability of C. difficile. I found that C. difficile is most prevalent (up to 76%) and abundant in healthy infants, and that C. difficile in infants is associated with multiple indicators of a healthy infant gut microbiome development. This study represents the largest single-species metagenomic survey to date, providing systematic, world-wide analysis of C. difficile carriage and associated microbiome composition.
Relevance of the study: Disentangling the real burden of C. difficile from other pathogenic species inducing the same symptomatology will help directing future research focus and fundings towards better diagnostic systems and diagnosis. In addition, understanding how C. difficile carriage remains asymptomatic will help understanding the mechnanisms behind CDI manifestation and progression.
SPIRE: a Searchable, Planetary-scale mIcrobiome REsource

Schmidt and Fullam, Ferretti et al. 2023 - Nucleic Acids Research
Metagenomic data in public repositories is continuously growing. While this represents an unprecedented opportunity to find global patterns in microbiome data, it also comes with challenges in processing such large amount of data and organize the associated metadata in a consistent, searchable manner. SPIRE provides ~16Tbp of metagenomic assemblies (contigs), 35+ billion genes predicted from those contigs, and over 1 million metagenome-assembled genomes (MAGs) of medium or high quality, obtained from ~100k metagenomic samples from >700 published studies. In this work, I heavily contributed to the datasets processing and metadata curation. The data can be accessed and downloaded at https://spire.embl.de/.
Relevance of the study: Building metagenome-assembled genomes (MAGs) from metagenomic data remains a time-consuming and computationally very intensive task. For these reasons, t is often prohibitive for most labs to generate and process MAGs for large datasets. SPIRE addresses that barrier by providing pre-computed MAGs, greatly accelerating meta-analyses and enabling more accessible, genome-resolved microbiome research.
METALOG: Curated and harmonised metadata for metagenomic data

Kuhn et al. 2025 - Nucleic Acids Research
As part of my research, I contributed to the development of a manually curated, global repository of metadata for metagenomic samples. With the growing scale of metagenomic datasets and meta-analyses, access to harmonized and standardized metadata remains a limiting factor. This resource addresses that need, providing curated metadata for over 110k metagenomic samples (~73k human samples, ~10k animal samples, ~5k ocean water samples, and 21k from other environments). In addition to metadata, Metalog offers pre-computed mOTUs3.0 and MetaPhlAn4.0 taxonomic profiles for rapid data exploration and integrates with the SPIRE database for genome-based analyses. More details in https://metalog.embl.de/.
Relevance of the study: Metadata for metagenomic studies are often scattered across multiple sources (e.g. publication text, supplementary tables, figures, public repositories), incomplete or erronoeous. METALOG addresses this issue by providing detailed, manually curated, and harmonized metadata for over 100,000 samples collected worldwide. When used in combination, METALOG and SPIRE offer a powerful framework that enables researchers to conduct robust microbiome studies and large-scale meta-analyses in a highly time- and resource-efficient manner.